Microbial Cell Factory Research Team
- About
- Services
- Highlights
- Research Personnel
- Contact
Microbial Cell Factory Research Team
Microbial Cell Factory Research Team aims to research and develop microorganisms, especially bacteria and yeasts, as effective hosts for improved heterologous expression of bioactive compounds by using modern biotechnology techniques and bioprocess development. Currently, our research team focuses on research and developments in 3 main areas.
1) Development of expression systems for recombinant proteins production by bacteria and yeasts
2) Synthetic biology technology for production of high-value chemicals and desired biofuels
3) Bioprocess development technology
1). Development of expression systems for recombinant proteins production by bacteria and yeasts
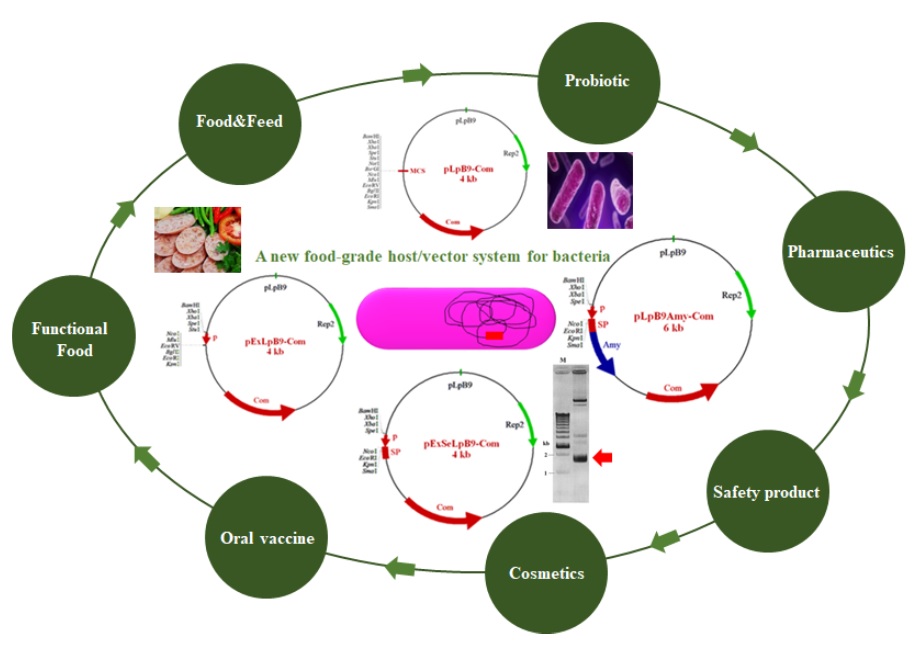
1.1 Construction of a new food-grade host/vector system for bacteria Heterologous gene expression in Lactobacillus plantarum or Bacillus subtilis has gained much attraction due to its high safety profile (Generally Recognized as Safe by US Food and Drug Administration) and potential probiotic properties. In addition, its genome data and relevant genetic tools are available. Fermentation technology of the bacterium has also been well established. Therefore, the development of a new food-grade host/vector system for bacteria is of highly interested. Previously, a series of plasmid vectors such as cloning, expression and secretion vectors were constructed based on native cryptic plasmid, pLpB9, originated from Nham starter culture, L. plantarum BCC9546. These shuttle vectors could replicate in L. plantarum, Bacillus subtilis and Escherichia coli. For expression vectors, both constitutive and inducible expression systems were constructed. A new salt-inducible expression vector was developed for the first time, as food-grade expression system. For secretion vectors, 12 novel secretion vectors were developed by applying 12 signal peptide sequences of L. plantarum into the expression vector. The function of these vectors was examined by expressing either GFP or some enzymes. High level of protein products were obtained from each recombinant plasmid in its appropriate hosts. All of these vectors are deposited at TBRC (Thailand Bioresource Research Center) and available for widely use in recombinant DNA technology. Nevertheless, since these vectors contain antibiotic resistance gene as selection marker, there is still safety concerns regarding dissemination of antibiotic resistance gene. To avoid this problem, the food- grade host/vector system has been considered as an alternative system. The system will allow broad industrial applications even in food, pharmaceutical and medical industries. Currently, the food-grade host/vector system of bacteria is being developed in our laboratory.
References
Promchai R., Boonchalearnb A., Visessanguanb W., Luxananil P. (2018) Rapid production of extracellular thermostable alkaline halophilic protease originating from an extreme haloarchaeon, Halobacterium salinarum by recombinant Bacillus subtilis. Biocat. Agri. Biotechnol. 15:192–198. Promchai R., Promdonkoy B., Tanapongpipat S., Visessanguan W., Eurwilaichitr L., Luxananil P. 2016. A novel salt-inducible vector for efficient expression and secretion of heterologous proteins in Bacillus subtilis. J Biotechnol. 222:86-93.
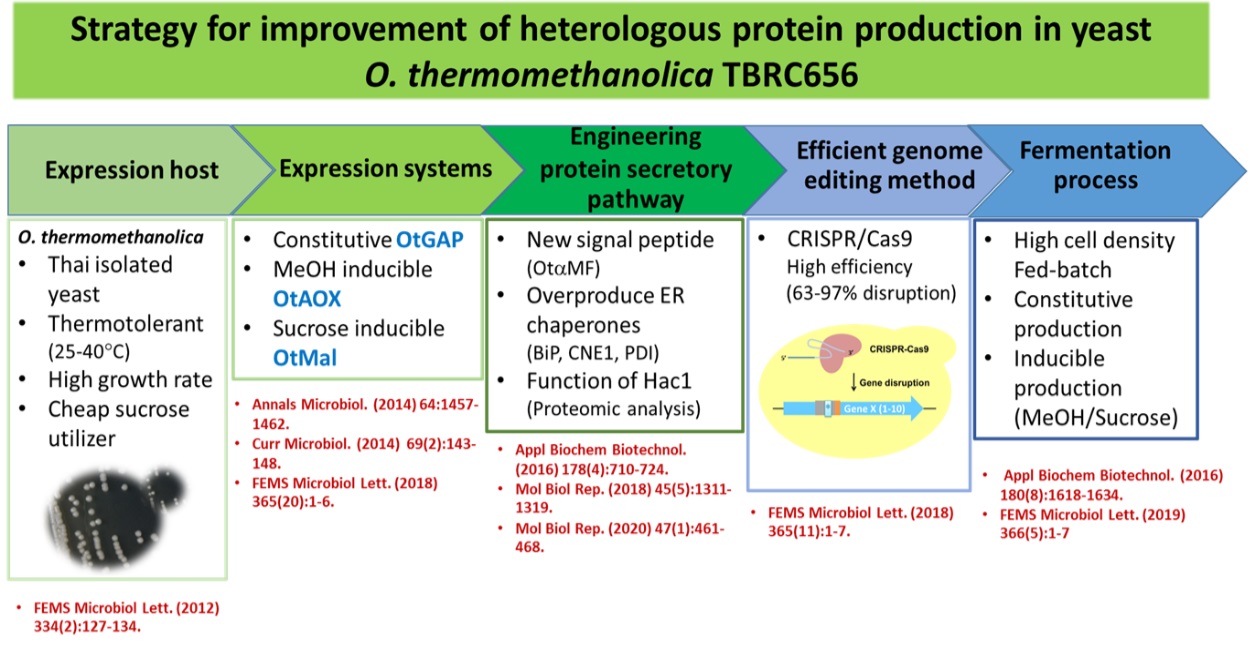
1.2 Development of thermotolerant yeast expression system for efficient protein production We have developed new expression system for recombinant protein production by Thai-isolated yeast Ogataea thermomethanolica TBRC656. O. thermomethanolica is thermotolerant yeast with its ability to utilize sucrose or molasses which is cheap and renewable carbon sources. These properties make it suitable for industrial application. To develop O. thermomethanolica as an efficient host, the potential of this yeast was initially demonstrated by its ability to be transformed at fairly high efficiency and perform N-glycosylation similar to that of Pichia pastoris. New expression vector was constructed such that it contains elements originated from O. thermomethanolica. Three native strong constitutive glyceraldehyde-3-phosphate dehydrogenease (GAP), methanol inducible alcohol oxidase (AOX) and sucrose inducible maltase (Mal) promoters were isolated and employed for protein production. Among
these 3 promoters, Mal promoter is more efficient to produce heterologous proteins from sucrose in yeast O. thermomethanolica than AOX and GAP promoters. To improve protein secretion, we explored the use of a native signal peptide sequence for directing heterologous protein secretion and overexpression of native ER-resident chaperone genes. The native signal peptide sequence was shown to promote higher secretion of heterologous proteins comparing with the classical signal peptide sequence of Saccharomyces cerevisiae. Overexpression of ER-resident chaperones also improved protein secretion depending on heterologous protein. To evaluate the feasibility of O. thermomethanolica as heterologous host in large-scale fermentation, the cost-effective media and fermentation process using sucrose (table sugar) as a sole carbon source was developed. These observations reveal the benefits of employing O. thermomethanolica as a host for heterologous protein production.
References
Phithakrotchanakoon C, Phaonakrop N, Roytrakul S, Tanapongpipat S, Roongsawang N. (2020) Protein secretion in wild-type and Othac1 mutant strains of thermotolerant methylotrophic yeast Ogataea thermomethanolica TBRC656. Mol Biol Rep. 47(1):461-468.
Boonchoo K., Puseenam A., Kocharin K., Tanapongpipat S., Roongsawang N. (2019) Sucrose-inducible heterologous expression of phytase in high cell density cultivation of the thermotolerant methylotrophic yeast Ogataea thermomethanolica. FEMS Microbiol Lett. 366(5). doi: 10.1093/femsle/fnz052.
Puseenam A., Kocharin K., Tanapongpipat S., Eurwilaichitr L., Ingsriswang S., Roongsawang N. (2018). A novel sucrose-based expression system for heterologous proteins expression in thermotolerant methylotrophic yeast Ogataea thermomethanolica. FEMS Microbiol Lett. 365(20). doi: 10.1093/femsle/fny238.
Phithakrotchanakoon C., Puseenam A., Phaonakrop N., Roytrakul S., Tanapongpipat S., Roongsawang N. (2018) Hac1 function revealed by the protein expression profile of a OtHAC1 mutant of thermotolerant methylotrophic yeast Ogataea thermomethanolica. Mol Biol Rep. 45(5):1311-1319.
Phithakrotchanakoon C., Puseenam A., Wongwisansri S., Eurwilaichitr L., Ingsriswang S., Tanapongpipat S., Roongsawang N. (2018) CRISPR-Cas9 enabled targeted mutagenesis in the thermotolerant methylotrophic yeast Ogataea thermomethanolica. FEMS Microbiol Lett. 365(11). doi: 10.1093/femsle/fny105.
Charoenrat T., Antimanon S., Kocharin K., Tanapongpipat S., Roongsawang N. (2016) High cell density process for constitutive production of a recombinant phytase in thermotolerant methylotrophic yeast Ogataea thermomethanolica using table sugar as carbon source. Appl Biochem Biotechnol. 180(8):1618-1634.
Roongsawang N., Puseenam A., Kitikhun S., Sae-Tang K., Harnpicharnchai P., Ohashi T., Fujiyama K., Tirasophon W., Tanapongpipat S. (2016) A novel potential signal peptide sequence and overexpression of ER-resident chaperones enhance heterologous protein secretion in thermotolerant methylotrophic yeast Ogataea thermomethanolica. Appl Biochem Biotechnol. 178(4):710-724.
Promdonkoy P., Tirasophon W., Roongsawang N., Eurwilaichitr L., Tanapongpipat S. (2014) Methanol- inducible promoter of thermotolerant methylotrophic yeast Ogataea thermomethanolica BCC16875 potential for production of heterologous protein at high temperatures. Curr Microbiol. 69(2):143-148.
Harnpicharnchai P., Promdonkoy P., Sae-Tang K., Roongsawang N., Tanapongpipat S. (2014) Use of the glyceraldehyde-3-phosphate dehydrogenase promoter from a thermotolerant yeast, Pichia thermomethanolica, for heterologous gene expression, especially at elevated temperature. Annals Microbiol. 64:1457-1462.
Tanapongpipat S., Promdonkoy P., Watanabe T., Tirasophon W., Roongsawang N., Chiba Y., Eurwilaichitr L. (2012) Heterologous protein expression in Pichia thermomethanolica BCC16875, a thermotolerant methylotrophic yeast and characterization of N-linked glycosylation in secreted protein. FEMS Microbiol Lett. 334(2):127-134.
2) Synthetic biology technology for production of high-value chemicals and desired biofuels
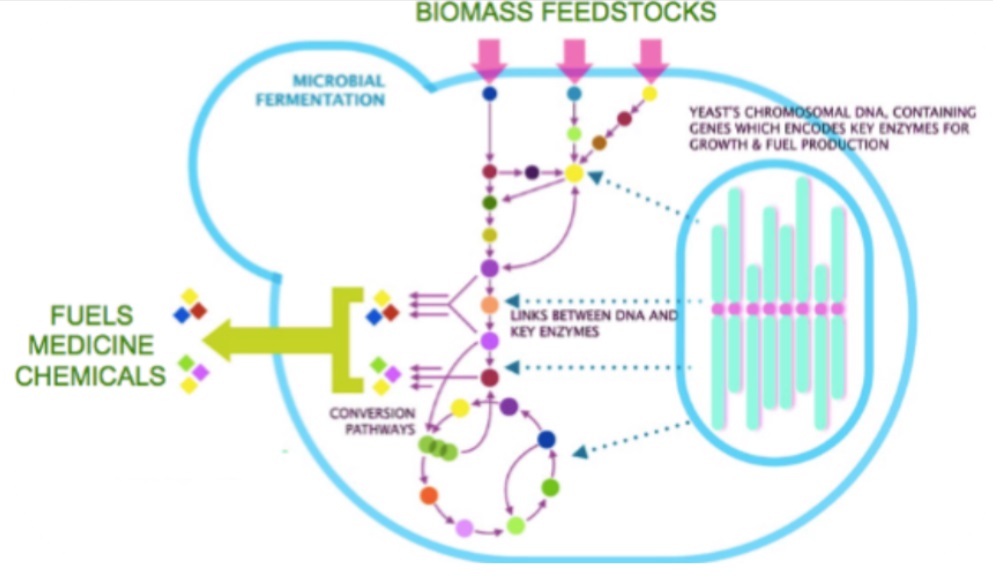
A major focus of our research team is the creation of efficient cell factories for converting plant biomass and other renewable feedstocks into desired biofuels and high-value chemicals. Our first aim is divided into three specific chemical targets: i) branched- and short-chain alcohols (biofuel replacement), ii) D-lactic acid (functional plastic monomer), and iii) carotenoids (food and feed/cosmetics ingredient). Our hosts of choice are the budding yeast Saccharomyces cerevisiae and the methylotrophic yeast Pichia pastoris. Moreover, we are creating biomass-tailored consolidated bioprocessing platforms (CBP) that can efficiently convert sugarcane bagasse—one of the most abundant agricultural wastes in Thailand—into the advanced biofuel isobutanol. For the second aim, we seek to optimize these metabolic pathways by employing a “synthetic biology toolbox” to fine-tune enzyme activity and expression levels, as well as minimize the build-up of toxic intermediates. Examples of these tools are gene circuit design, CRISPR-based genome engineering, compartmentalization of pathway enzymes. Our choice of utilizing the yeasts S. cerevisiae and P. pastoris as eukaryotic models to develop synthetic biology tools offers many advantages, such as the availability of multi-omics database and well established genetic tools, ease of maintenance and a proven track record in various industrial applications. We envision that our synthetic biology platform will pave the way for a scalable, controllable and economical route to biofuel and chemical production.
References
Siripong W., Angela C.K., Tanapongpipat S., Runguphan W. (2020) Metabolic Engineering of Pichia
pastoris for production of isopentanol (3-methyl-1-butanol) (In press)
Promdonkoy P., Siripong W., Downes J.J., Tanapongpipat S., Runguphan, W. (2019) Systematic
improvement of isobutanol production from D-xylose in engineered Saccharomyces cerevisiae.
AMB Express 9:160.
Siripong W., Wolf P., Putri T., Kocharin K., Tanapongpipat S., Runguphan W. (2018) Metabolic engineering
of the methylotrophic yeast Pichia pastoris for production of isobutanol and isobutyl acetate.
Biotechnol Biofuels. 11:1.
3) Bioprocess development technology
Bioprocess engineering is a specialization field of study including biotechnology, biological engineering,
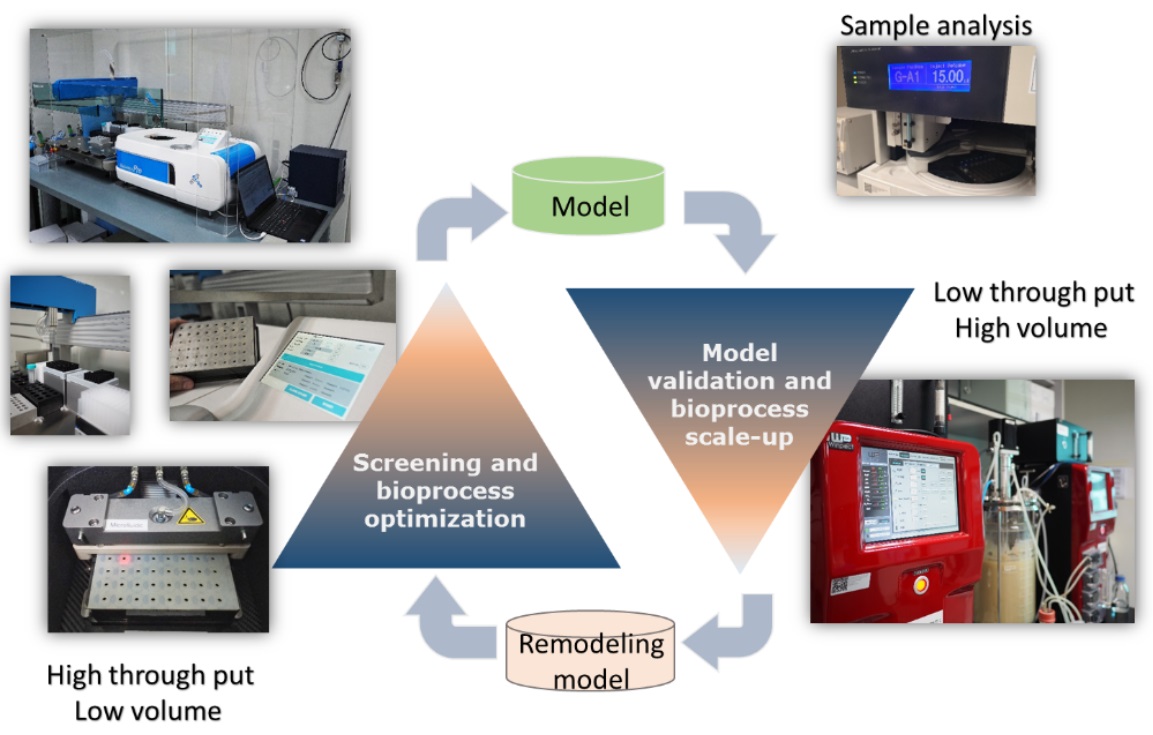
chemical engineering and of agricultural engineering. This research field deals with the design and development of equipment and processes both upstream and downstream by employing the living organisms and their components as a cell factory for the manufacturing of industrial biological products. At present, many research carried out by Microbial Cell Factory Research Team is in a transition phase to bring the biological products from a bench-scale production in the laboratory to the market. In order to execute a process scale-up, an efficient bioprocess optimization using Design of Experiment (DOEs) together with a validation of bioprocessing model are required. The main technological objectives of bioprocess optimization carried out in our research team include process scale-up with reducing cost of production by using low-cost media, improve the productivity by shorten the fermentation time as well as develop a consistency bioprocess that can be scalable. Recent technology established at BIOTEC include high-throughput fermentations with the online monitoring of fermentation parameters e.g. biomass, pH and DO along with the appropriate sample analysis tools will facilitate and ensure a successful transition from lab scale to commercial scale production of the market desired biological products. Our currently research activity on bioprocess scale-up and optimization is focusing on the production of stereospecific D-lactic acid to function as a plastic monomer, carotenoids as anti-oxidative compound and feed enzyme for poultry industry.
1. Bacterial expression system
All vectors are deposited at TBRC (Thailand Bioresource Research Center) and available for widely use in recombinant DNA technology.
Relevant IPs
Patent: Plasmid for halophilic protease secretion by B. subtilis. Publication number 1601006067.
Patent: Plasmid vectors for secreted protein expression in Lactobacillus plantarum, Bacillus subtilis and Escherichia coli. Publication number 1601004946.
Patent : Salt-inducible plasmid for protein expression in Bacillus subtilis. Publication number 1401004416.
Petty patent : Plasmid vectors for secreted protein expression in Bacillus subtilis. Publication number 1603001560.
Petty patent: Plasmid vectors for secreted protein expression in Lactobacillus plantarum and Bacillus subtilis. Publication number 1603001559.
Petty patent: Cloning plasmid of bacteria. Petty patent number 10415.
2. OTExpress : High efficient and low cost protein expression system by thermotolerant yeast
Protein production by thermotolerant yeast is high efficient and low cost system. This thermotolerant yeast is a potential heterologous host which is capable for expressing heterologous protein over a wide range of temperature and can utilize various carbon sources including sucrose and sugarcane molasses which is cheap and renewable carbon sources.
Relevant IPs
Petty patent: Bioprocess for production heterologous protein from sucrose by engineered thermotolerant yeast Ogataea thermomethanolica. Publication number 1803002676.
Petty patent: Products for expression of heterologous proteins from sucrose and methods of making thereof. Publication number 1803001465.
Petty patent: Products for methanol-inducible gene expression to produce target proteins or biological compounds from thermotolerant yeast and the process of using such products. Publication number 1703001117.
Petty patent: Products for constitutive gene expression to produce target proteins or biological compounds from thermotolerant yeast and the process of using such products. Publication number 1703001116.
Petty patent: Recombinant plasmid for extracellular expression of heterologous protein from sucrose in yeast Ogataea spp. Petty patent number 14258.
Petty patent: Bioprocess for constitutive production of heterologous protein by thermotolerant yeast Ogataea thermomethanolica. Petty patent number 14019.
Petty patent: Recombinant plasmid for extracellular expression of heterologous protein in yeast Ogataea spp. Petty patent number 10661.
3. Microbial production of biofuels and high-value chemicals
We applied synthetic biology and metabolic engineering tools to engineer industrially relevant microbes to produce biofuels and high-value chemicals from renewable carbon sources. Our patented engineered microbes are ready for pre-commercial scale testing. Our inventions have been recognized at the national and international levels.
Relevant IPs
Patent: An episomal plasmid system for high level gene expression. Publication number 1701005536.
Petty patent: Recombinant Saccharomyces cerevisiae (BMGC330) for high-level production of isoprenoid compounds. Publication number 1903002092.
Petty patent: Recombinant Saccharomyces cerevisiae (BMGC330) for isobutanol production from xylose or xylose-containing biomass. Publication number 1903002093.
Petty patent: Recombinant Pichia pastoris for isopentanol (3-methyl-1-butanol) production. Publication number 1803002193.
Petty patent: Recombinant Saccharomyces cerevisiae for production of fragrant terpenoids. Publication number 1803002245.
Petty patent: Recombinant Pichia pastoris for isobutanol production. Publication number 1703001862.
Petty patent: Recombinant Pichia pastoris for production of fragrant ester of branched-chain alcohols. Publication number 1703001861.
Petty patent: CRISPR-dCas9 system for fine-tuning carotenoid pathway in recombinant yeasts. Publication number 1603002635.
Petty patent: Development of a CRISPR-dCas9 system for fine-tuning metabolic pathways in recombinant microorganisms. Publication number 1603002634.
4. Bioprocess
Our research team has developed the bioprocess for industrial enzyme production e.g. feed enzyme, biomass hydrolysis enzyme. Recently, the developed technology on process scale-up of these products has been transferred to the industrial partnership for trial production in a pilot scale.
Relevant IPs
Patent: Formulation process of xylanase with additives for enhancing catalytic activity of the enzyme. Publication number 1701003741.
Petty patent: Culture media formulation and the fermentation process using thereof specified for the production of feed enzyme. Publication number 1803001776.
Petty patent: Fermentation process for high cell density cultivation of bacteria without oxygen enrichment for the production of enzyme digesting plant biomass. Publication number 1703001161.
Microbial Cell Factory Research Team


Sutipa Tanapongpipat
Research Team Leader
Principal researcher

Plearnpis Luxananil
Researcher

Weerawat Runguphan
Researcher

Akaraphol Watcharawipas
Postdoctoral researcher

Worarat Kruasuwan
Postdoctoral researcher

Peerada Promdonkoy
Research assistant

Kittapong Sae-Tang
Senior Research assistant

Ruangurai Promchai
Research assistant

Aekkachai Puseenam
Research assistant

Kitisak Sansatchanon
Research assistant

Pipat Sudying
Co-research assistant

Pornsiri Bumrungtham
Co-research assistant
Contact
Microbial Cell Factory Research Team
Dr. Sutipa Tanapongpipat
National Center for Genetic Engineering and Biotechnology ( BIOTEC)
National Science and Technology Development Agency (NSTDA)
113 Thailand Science Park
Phahonyothin Road
Khlong Nueng, Khlong Luang
Pathum Thani 12120 Thailand
Tel: 66 2564 6700 Ext 3472
Fax: 66 2564 6707




